library(glue)
library(ggplot2)
library(bench)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, unionGlue is advertised as
Fast, dependency free string literals
So what do we mean when we say that glue is fast? This does not mean glue is the fastest thing to use in all cases, however for the features it provides we can confidently say it is fast.
A good way to determine this is to compare its speed of execution to some alternatives.
-
base::paste0(),base::sprintf(): Functions in base R implemented in C that provide variable insertion (but not interpolation). -
R.utils::gstring(): Provides a similar interface as glue, but uses${}to delimit blocks to interpolate. -
pystr::pystr_format()1,rprintf::rprintf(): Provide an interface similar to python string formatters with variable replacement, but not arbitrary interpolation.
Note: stringr::str_interp() was previously included in
this benchmark, but is now formally marked as “superseded”, in favor of
stringr::str_glue(), which just calls
glue::glue().
Simple concatenation
bar <- "baz"
simple <- bench::mark(
glue = as.character(glue::glue("foo{bar}")),
gstring = R.utils::gstring("foo${bar}"),
paste0 = paste0("foo", bar),
sprintf = sprintf("foo%s", bar),
rprintf = rprintf::rprintf("foo$bar", bar = bar)
)
simple %>%
select(expression:total_time) %>%
arrange(median)
#> # A tibble: 5 × 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 sprintf 731.09ns 802.1ns 1153386. 0B 0
#> 2 paste0 1.45µs 1.62µs 587959. 0B 0
#> 3 glue 94.52µs 101.24µs 9611. 141.56KB 25.0
#> 4 gstring 218.74µs 231.9µs 4199. 2.45MB 16.9
#> 5 rprintf 274.46µs 285.39µs 3446. 503.39KB 8.23
# plotting function defined in a hidden chunk
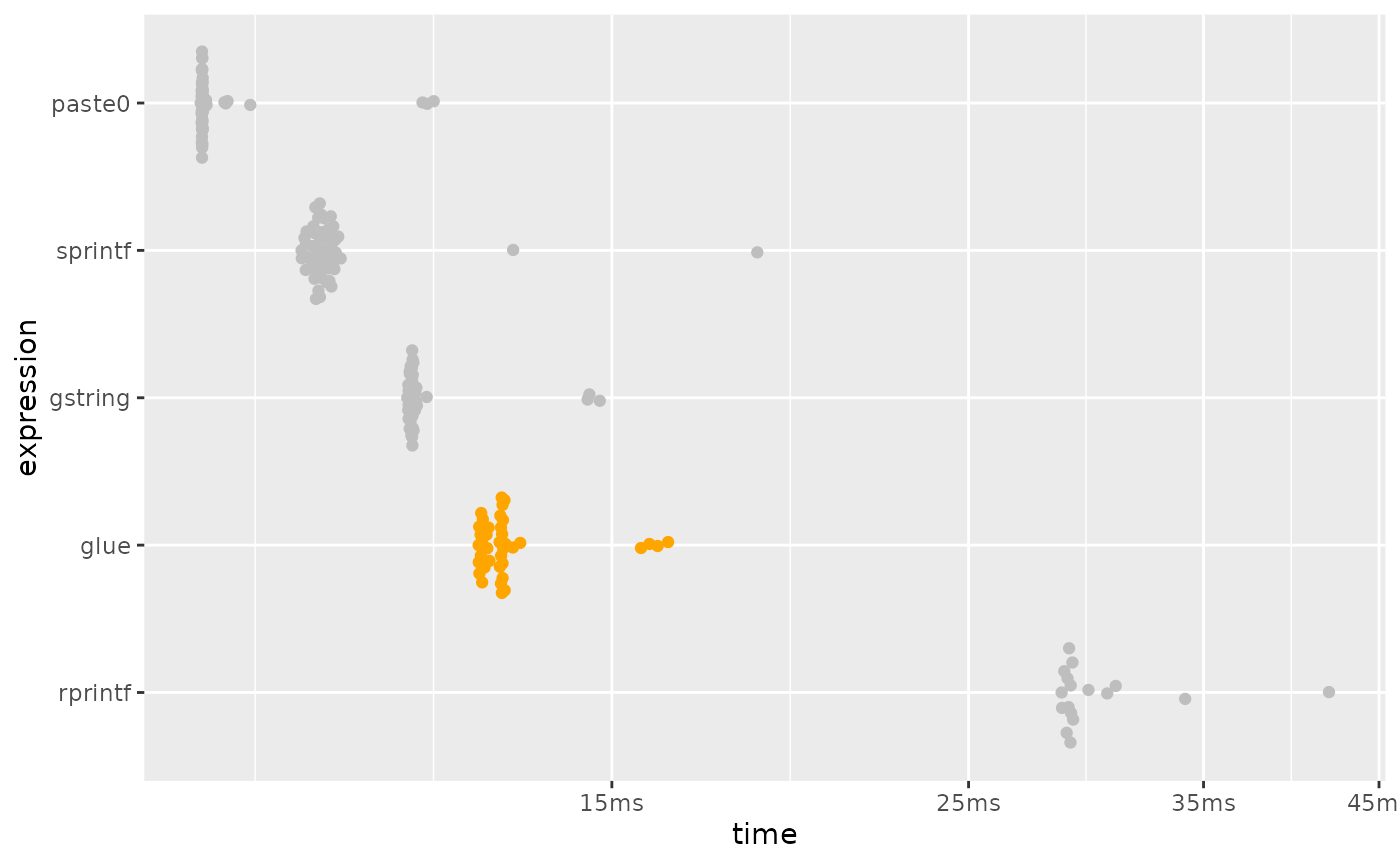
plot_comparison(simple)
#> Warning: The `trans` argument of `continuous_scale()` is deprecated as of ggplot2
#> 3.5.0.
#> ℹ Please use the `transform` argument instead.
#> This warning is displayed once every 8 hours.
#> Call `lifecycle::last_lifecycle_warnings()` to see where this warning was
#> generated.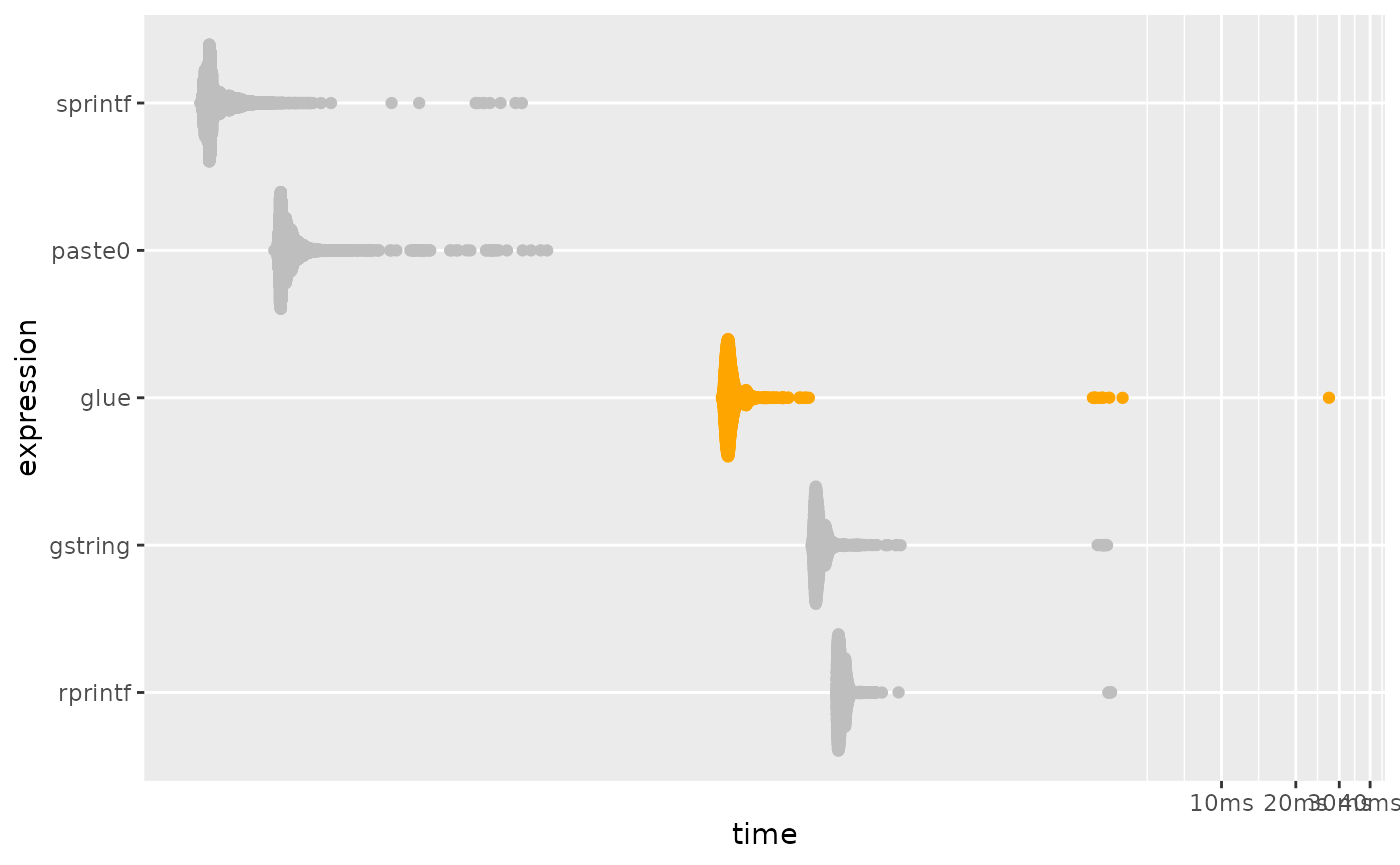
While glue() is slower than paste0 and
sprintf(), it is twice as fast as gstring(),
and rprintf().
Although paste0() and sprintf() don’t do
string interpolation and will likely always be significantly faster than
glue, glue was never meant to be a direct replacement for them.
rprintf::rprintf() does only variable interpolation, not
arbitrary expressions, which was one of the explicit goals of writing
glue.
So glue is ~2x as fast as the function (gstring()),
which has roughly equivalent functionality.
It also is still quite fast, with over 8000 evaluations per second on this machine.
Vectorized performance
Taking advantage of glue’s vectorization is the best way to improve
performance. In a vectorized form of the previous benchmark, glue’s
performance is much closer to that of paste0() and
sprintf().
bar <- rep("bar", 1e5)
vectorized <- bench::mark(
glue = as.character(glue::glue("foo{bar}")),
gstring = R.utils::gstring("foo${bar}"),
paste0 = paste0("foo", bar),
sprintf = sprintf("foo%s", bar),
rprintf = rprintf::rprintf("foo$bar", bar = bar)
)
vectorized %>%
select(expression:total_time) %>%
arrange(median)
#> # A tibble: 5 × 6
#> expression min median `itr/sec` mem_alloc `gc/sec`
#> <bch:expr> <bch:tm> <bch:tm> <dbl> <bch:byt> <dbl>
#> 1 paste0 8.25ms 8.27ms 119. 781.3KB 6.36
#> 2 sprintf 8.96ms 9ms 111. 781.3KB 6.40
#> 3 gstring 11.09ms 11.14ms 89.7 1.53MB 8.97
#> 4 glue 11.27ms 11.88ms 83.2 2.29MB 11.6
#> 5 rprintf 27.45ms 27.65ms 36.1 3.05MB 5.56
# plotting function defined in a hidden chunk
plot_comparison(vectorized)